Keywords: DC resistivity, 1D sounding, inversion, parametric, sparse norm, apparent resistivity, wires mapping.
Summary: Here we invert DC resistivity data for a single Wenner sounding. We demonstrate 3 approaches that can be used to invert 1D data in SimPEG:
A weighted least-squares inversion where the number of layers and their thicknesses are fixed.
An iteratively reweighted least-squares (IRLS) inversion to recover sparse and/or blocky structures.
A parametric inversion where we invert for the layer thicknesses and electrical properties assuming a 3-layered Earth.
The weighted least-squares approach is a great introduction to geophysical inversion with SimPEG. One drawback however, is that it recovers smooth structures which may not be representative of the true model. To recover sparse and/or blocky 1D structures, we demonstrate an iteratively reweighted least-squares approach. If the number of layers is known, but their depths, thicknesses and conductivities are not, we can use a parametric inversion approach.
Learning Objectives: Because this tutorial focusses primarily on inversion-related functionality, we urge the reader to become familiar with functionality explained in the 1D Forward Simulation of DC Resistivity Data tutorial before working through this one. For this tutorial, we focus on:
General approaches for 1D inversion with SimPEG.
How to assign appropriate uncertainties to apparent resistivity or normalized voltage data.
Choosing suitable parameters for the inversion.
Specifying directives that are applied throughout the inversion.
Weighted least-squares, sparse-norm and parametric inversion.
Analyzing inversion outputs.
Import Modules¶
Here, we import all of the functionality required to run the notebook for the tutorial exercise.
All of the functionality specific to the forward simulation of 1D DC resistivity data are imported from the simpeg
# SimPEG functionality
from simpeg.electromagnetics.static import resistivity as dc
from simpeg.utils import plot_1d_layer_model, download
from simpeg import (
maps,
data,
data_misfit,
regularization,
optimization,
inverse_problem,
inversion,
directives,
)
# discretize functionality
from discretize import TensorMesh
# Basic Python functionality
import os
import numpy as np
import matplotlib as mpl
import matplotlib.pyplot as plt
import tarfile
mpl.rcParams.update({"font.size": 14})Download Tutorial Files¶
For this tutorial, the observed data for the Wenner sounding are stored within a tar-file. Here, we download and extract the data file.
# URL to download from repository assets
data_source = "https://github.com/simpeg/user-tutorials/raw/main/assets/05-dcr/inv_dcr_1d_files.tar.gz"
# download the data
downloaded_data = download(data_source, overwrite=True)
# unzip the tarfile
tar = tarfile.open(downloaded_data, "r")
tar.extractall()
tar.close()
# path to the directory containing our data
dir_path = downloaded_data.split(".")[0] + os.path.sep
# files to work with
data_filename = dir_path + "app_res_1d_data.dobs"overwriting /home/ssoler/git/user-tutorials/notebooks/05-dcr/inv_dcr_1d_files.tar.gz
Downloading https://github.com/simpeg/user-tutorials/raw/main/assets/05-dcr/inv_dcr_1d_files.tar.gz
saved to: /home/ssoler/git/user-tutorials/notebooks/05-dcr/inv_dcr_1d_files.tar.gz
Download completed!
/tmp/ipykernel_843544/329559176.py:9: DeprecationWarning: Python 3.14 will, by default, filter extracted tar archives and reject files or modify their metadata. Use the filter argument to control this behavior.
tar.extractall()
Load and Plot the Data¶
Here, we load and parse the data file containing observed data and survey geometry. The data file contains the xyz locations for the A, B, M and N electrodes, in order, as well as the observed data. We then compute the AB/2 (half separation distance) and plot the Wenner sounding data.
# Load data
dobs = np.loadtxt(str(data_filename))# Extract A, B, M and N electrode locations and the observed data
A_electrodes = dobs[:, 0:3]
B_electrodes = dobs[:, 3:6]
M_electrodes = dobs[:, 6:9]
N_electrodes = dobs[:, 9:12]
dobs = dobs[:, -1]# Compute the AB/2 separation for the 1D Wenner array
AB_separations = np.sqrt(np.sum((A_electrodes - B_electrodes) ** 2, axis=1))# Plot apparent resistivity as a function of AB/2
fig = plt.figure(figsize=(8, 4))
mpl.rcParams.update({"font.size": 14})
ax1 = fig.add_axes([0.15, 0.1, 0.7, 0.85])
ax1.semilogy(AB_separations / 2, dobs, "b-o")
ax1.grid(which="both")
ax1.set_xlabel("AB/2 (m)")
ax1.set_ylabel(r"Apparent Resistivity ($\Omega m$)")
plt.show()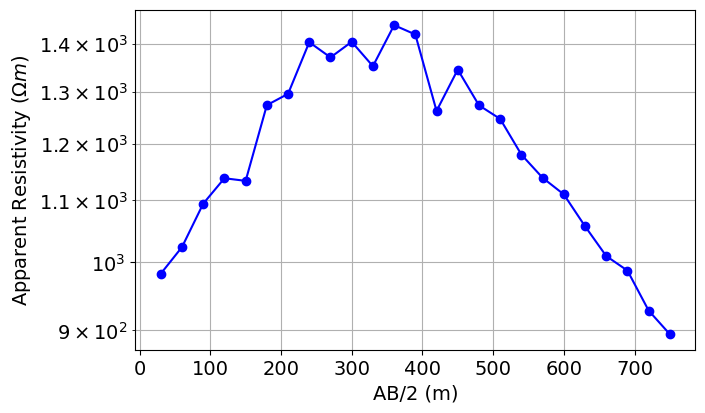
Assign Uncertainties¶
Inversion with SimPEG requires that we define the uncertainties on our data; that is, an estimate of the standard deviation of the noise on our data assuming it is uncorrelated Gaussian with zero mean. An online resource explaining uncertainties and their role in the inversion can be found here.
For 1D apparent resistivity data, we generally apply a percent uncertainty to all data. A percent uncertainty will fit conductive and resistive structures equally; unlike a floor uncertainty which will prioritize fitting more resistive structures. For this tutorial, we apply a 2% uncertainty to all data. Depending on the quality of the data, we may choose a percent uncertainty between 2% - 10%.
For 1D normalized voltage data, we also tend to apply a percent uncertainty to all data. Differences in electrode spacing and subsurface conductivity result in measured voltages that span many orders of magnitude. A percent uncertainty ensures all data are fit equally. Like apparent resistivity, a percent uncertainty of 2% - 10% is a good initial guess.
NOTE: if the data contain very small values that you feel could be erroneous, a small floor value can be added to ensure stability of the inversion.
# Add 2.5% uncertainties to all data.
uncertainties = 0.025 * np.abs(dobs)Define the Survey¶
Here, we define the survey geometry. Although the survey consists of a single 1D Wenner sounding, the approach is generalized for 2D and 3D surveys where the electrode locations may not be sorted. For a comprehensive description of constructing DC resistivity surveys in SimPEG, see the 1D Forward Simulation of DC Resistivity Data tutorial.
# Sort by unique current electrode locations
unique_tx, k = np.unique(np.c_[A_electrodes, B_electrodes], axis=0, return_index=True)
n_sources = len(k)
k = np.sort(k)
k = np.r_[k, len(k) + 1]# Define empty list for sources to live
source_list = []
# Loop over all sources
for ii in range(0, n_sources):
# MN electrode locations for receivers. Each is an (N, 3) numpy array
M_locations = M_electrodes[k[ii] : k[ii + 1], :]
N_locations = N_electrodes[k[ii] : k[ii + 1], :]
receiver_list = [
dc.receivers.Dipole(
M_locations,
N_locations,
data_type="apparent_resistivity",
)
]
# AB electrode locations for source. Each is a (1, 3) numpy array
A_location = A_electrodes[k[ii], :]
B_location = B_electrodes[k[ii], :]
source_list.append(dc.sources.Dipole(receiver_list, A_location, B_location))
# Define survey
survey = dc.Survey(source_list)Define the Data¶
The SimPEG Data class is required for inversion and connects the observed data, uncertainties and survey geometry.
data_object = data.Data(survey, dobs=dobs, standard_deviation=uncertainties)Weighted Least-Squares Inversion¶
Here, we use the weighted least-squares inversion approach to recover the log-conductivities on a 1D layered Earth. We impose no a-priori information about the number of layers (geological units) or their thicknesses. Instead, we define a large number of layers with exponentially increasing thicknesses. And the depth, thickness and electrical properties of the Earth are inferred from the recovered model.
Design a 1D Layered Earth¶
When the spacings between the electrodes are small, we are obtaining higher resolution information about the conductivity near the Earth’s surface. As a result, we define much thinner layers near the Earth’s surface. When the spacings between electrodes are large, we are obtaining lower resolution information about the conductivity at depth. So at depth, we can define layers that are much thicker. The thickness defining the top layer is ultimately determined by the minimum electrode spacing. And the thickness and depth to the lower layer is determined by the largest electrode spacing. The rate at which the layer thicknesses increase with depth is determined by the user.
For Wenner soundings: For the 1D Wenner array, the layer thicknesses and largest depth are determined by the AB/2 spacing. A reasonable minimum layer thickness is 10% the minimum AB/2 spacing. And the depth to the bottom layer should be roughly equal to the largest AB/2 spacing.
For dipole-dipole, pole-dipole, etc...: For these surveys, the source electrodes AB and receiver electrodes MN are offset from one another. For a particular set of electrodes ABMN, we consider the distance between the average source location and average receiver location. For a dipole-dipole survey, this would be:
For a pole-dipole survey, this would be:
Pole sources penetrate the Earth more deeply and produce smoother fields within the source region, so we can get away with a larger layer thickness for the top layer but we need to discretize deeper. But in general, it is easy to make your your top layer equal to 10% of your smallest distance. And to make the depth to the lower layer equal to the largest distance.
# Use Wenner electrode spacings to set discretization parameters
print("AB separations: {}".format(AB_separations))
depth_min = 5.0 # top layer thickness
depth_max = np.max(AB_separations / 2) # depth to lowest layer
geometric_factor = 1.1 # rate of thickness increaseAB separations: [ 60. 120. 180. 240. 300. 360. 420. 480. 540. 600. 660. 720.
780. 840. 900. 960. 1020. 1080. 1140. 1200. 1260. 1320. 1380. 1440.
1500.]
# Increase subsequent layer thicknesses by the geometric factors until
# it reaches the maximum layer depth.
layer_thicknesses = [depth_min]
while np.sum(layer_thicknesses) < depth_max:
layer_thicknesses.append(geometric_factor * layer_thicknesses[-1])
n_layers = len(layer_thicknesses) + 1 # Number of layersDefine a Model and Mapping¶
Recall from the Forward Simulation of 1D Sounding DC Resistivity Data tutorial that the ‘model’ is not necessarily synonymous with physical property values. And that we need to define a mapping from the model to the set of input parameters required by the forward simulation. When inverting to recover electrical resistivities (or conductivities), it is best to use the log-value, as electrical resistivities (or conductivities) of rocks span many order of magnitude.
Here, the model defines the log-resistivity values for a defined set of subsurface layers. And we use the simpeg.maps.ExpMap to map from the model parameters to the resistivity values required by the forward simulation.
log_resistivity_map = maps.ExpMap(nP=n_layers)Starting/Reference Models¶
The starting model defines a reasonable starting point for the inversion. Because electromagnetic problems are non-linear, your choice in starting model does have an impact on the recovered model. For DC resistivity inversion, we generally choose our starting model based on apparent resistivities. For the tutorial example, the apparent resistivities were near 1000 . It should be noted that the starting model cannot be vector of zeros, otherwise the inversion will be unable to compute a gradient direction at the first iteration.
The reference model is used to include a-priori information. The impact of the reference model on the inversion will be discussed in another tutorial. The reference model for basic inversion approaches is either zero or equal to the starting model.
Notice that the length of the starting and reference models is equal to the number of model parameters!!!
# Starting model is log-resistivity values (Ohmm)
starting_resistivity_model = np.log(1e3 * np.ones(n_layers))
# Reference model is also log-resistivity values (Ohmm)
reference_resistivity_model = starting_resistivity_model.copy()Define the Forward Simulation¶
A simulation object defining the forward problem is required in order to predict data and calculate misfits for recovered models. A comprehensive description of the simulation object for 1D DC resistivity was discussed in the 1D Forward Simulation of DC Resistivity Data for a Single Sounding tutorial. Here, we use the Simulation1DLayers which simulates the data according to a 1D Hankel transform solution.
The layer thicknesses are a static property of the simulation, and we set them using the thicknessess keyword argument. Since our model consists of log-resistivities, we use rhoMap to set the mapping from the model to the layer resistivities.
simulation_L2 = dc.simulation_1d.Simulation1DLayers(
survey=survey,
rhoMap=log_resistivity_map,
thicknesses=layer_thicknesses,
)Define the Data Misfit¶
To understand the role of the data misfit in the inversion, please visit this online resource. Here, we use the L2DataMisfit class to define the data misfit. In this case, the data misfit is the L2 norm of the weighted residual between the observed data and the data predicted for a given model. When instantiating the data misfit object within SimPEG, we must assign an appropriate data object and simulation object as properties.
dmis_L2 = data_misfit.L2DataMisfit(simulation=simulation_L2, data=data_object)Define the Regularization¶
To understand the role of the regularization in the inversion, please visit this online resource.
To define the regularization within SimPEG, we must define a 1D tensor mesh. Meshes are designed using the discretize package. Whereas layer thicknesses and our model are defined from our top-layer down, tensor meshes are defined from the bottom up. So to define a 1D tensor mesh for the regularization, we:
add an extra layer to the end of our thicknesses so that the number of cells in the 1D mesh equals the number of model parameters
reverse the order so that the model parameters in the regularization match up with the appropriate cell
define the tensor mesh from the cell widths
# Define 1D cell widths
h = np.r_[layer_thicknesses, layer_thicknesses[-1]]
h = np.flipud(h)
# Create regularization mesh
regularization_mesh = TensorMesh([h], "N")
print(regularization_mesh)
TensorMesh: 31 cells
MESH EXTENT CELL WIDTH FACTOR
dir nC min max min max max
--- --- --------------------------- ------------------ ------
x 31 -901.79 -0.00 5.00 79.32 1.10
By default, the regularization acts on the model parameters. In this case, the model parameters are the log-resistivities, not the electric resistivities!!! Here, we use the WeightedLeastSquares regularization class to constrain the inversion result. Here, length scale along x are used to balance the smallness and smoothness terms; yes, x is smoothness along the vertical direction. And the reference model is only applied to the smallness term. If we wanted to apply the regularization to a function of the model parameters, we would need to set an approprate mapping object using the mapping keyword argument.
reg_L2 = regularization.WeightedLeastSquares(
regularization_mesh,
length_scale_x=1.0,
reference_model=reference_resistivity_model,
reference_model_in_smooth=False,
)Define the Optimization Algorithm¶
Here, we use the InexactGaussNewton class to solve the optimization problem using the inexact Gauss Newton with conjugate gradient solver. Reasonable default values have generally been set for the properties of each optimization class. However, the user may choose to set custom values; e.g. the accuracy tolerance for the conjugate gradient solver or the number of line searches.
opt_L2 = optimization.InexactGaussNewton(
maxIter=100, maxIterLS=20, cg_maxiter=20, cg_rtol=1e-3
)Define the Inverse Problem¶
We use the BaseInvProblem class to fully define the inverse problem that is solved at each beta (trade-off parameter) iteration. The inverse problem requires appropriate data misfit, regularization and optimization objects.
inv_prob_L2 = inverse_problem.BaseInvProblem(dmis_L2, reg_L2, opt_L2)Provide Inversion Directives¶
Directives represent operations that are carried out during the inversion. Here, we apply common directives for weighted least-squares inversion of DC resistivity data and describe their roles. These are:
UpdateSensitivityWeights: Apply sensitivity weighting to counteract the natural tendency of DC resistivity inversion to place materials near the electrodes. Since the problem is non-linear and the sensitivities are updated with every model, we set
every_iteration=True.UpdatePreconditioner: Apply Jacobi preconditioner when solving optimization problem to reduce the number of conjugate gradient iterations. We set
update_every_iteration=Truebecause the ideal preconditioner is model-dependent.BetaEstimate_ByEig: Compute and set starting trade-off parameter (beta) based on largest eigenvalues.
BetaSchedule: Size reduction of the trade-off parameter at every beta iteration, and the number of Gauss-Newton iterations for each beta. In general, a
coolingFactorbetween 1.5 and 2.5, andcoolingRateof 2 or 3 works well for DC resistivity inversion. Cooling beta too quickly will result in portions of the model getting trapped in local minima. And we will not be finding the solution that minimizes the optimization problem if the cooling rate is too small.TargetMisfit: Terminates the inversion when the data misfit equals the target misfit. A
chifact=1terminates the inversion when the data misfit equals the number of data.
The directive objects are organized in a list. Upon starting the inversion or updating the recovered model at each iteration, the inversion will call each directive within the list in order. The order of the directives matters, and SimPEG will throw an error if directives are organized into an improper order. Some directives, like the BetaEstimate_ByEig are only used when starting the inversion. Other directives, like UpdatePreconditionner, are used whenever the model is updated.
sensitivity_weights = directives.UpdateSensitivityWeights(every_iteration=True)
update_jacobi = directives.UpdatePreconditioner(update_every_iteration=True)
starting_beta = directives.BetaEstimate_ByEig(beta0_ratio=5)
beta_schedule = directives.BetaSchedule(coolingFactor=2.0, coolingRate=2)
target_misfit = directives.TargetMisfit(chifact=1.0)
directives_list_L2 = [
sensitivity_weights,
update_jacobi,
starting_beta,
beta_schedule,
target_misfit,
]Define and Run the Inversion¶
We define the inversion using the BaseInversion class. The inversion class must be instantiated with an appropriate inverse problem object and directives list. The run method, along with a starting model, is respondible for running the inversion. The output is a 1D numpy.ndarray containing the recovered model parameters
# Here we combine the inverse problem and the set of directives
inv_L2 = inversion.BaseInversion(inv_prob_L2, directives_list_L2)
# Run the inversion
recovered_model_L2 = inv_L2.run(starting_resistivity_model)INFO: Directive TargetMisfit: Target data misfit is 25.0
Running inversion with SimPEG v0.25.0
============================ Inexact Gauss Newton ============================
# beta phi_d phi_m f |proj(x-g)-x| LS Comment
-----------------------------------------------------------------------------
0 5.17e+02 1.42e+03 0.00e+00 1.42e+03
1 5.17e+02 1.11e+03 2.64e-01 1.25e+03 1.88e+03 0
2 5.17e+02 1.10e+03 2.77e-01 1.25e+03 4.41e+01 0
3 2.59e+02 9.27e+02 7.72e-01 1.13e+03 7.77e+02 0
4 2.59e+02 9.23e+02 7.95e-01 1.13e+03 2.36e+01 0
5 1.29e+02 7.29e+02 1.86e+00 9.69e+02 6.62e+02 0
6 1.29e+02 7.26e+02 1.91e+00 9.73e+02 2.04e+01 0
7 6.47e+01 5.47e+02 3.86e+00 7.97e+02 5.18e+02 0
8 6.47e+01 5.47e+02 3.98e+00 8.05e+02 1.77e+01 0
9 3.23e+01 3.93e+02 7.36e+00 6.31e+02 3.78e+02 0
10 3.23e+01 3.99e+02 7.56e+00 6.43e+02 2.28e+01 0
11 1.62e+01 2.69e+02 1.32e+01 4.83e+02 2.63e+02 0
12 1.62e+01 2.80e+02 1.35e+01 4.99e+02 3.11e+01 0
13 8.08e+00 1.78e+02 2.25e+01 3.59e+02 1.80e+02 0
14 8.08e+00 1.92e+02 2.29e+01 3.77e+02 3.88e+01 0
15 4.04e+00 1.16e+02 3.59e+01 2.61e+02 1.22e+02 0
16 4.04e+00 1.30e+02 3.67e+01 2.78e+02 4.50e+01 0
17 2.02e+00 7.78e+01 5.43e+01 1.88e+02 8.12e+01 0
18 2.02e+00 8.92e+01 5.59e+01 2.02e+02 4.97e+01 0
19 1.01e+00 5.48e+01 7.85e+01 1.34e+02 5.31e+01 0
20 1.01e+00 6.32e+01 8.19e+01 1.46e+02 5.31e+01 0
21 5.05e-01 4.11e+01 1.10e+02 9.67e+01 3.36e+01 0
22 5.05e-01 4.65e+01 1.15e+02 1.05e+02 5.41e+01 0
23 2.53e-01 3.31e+01 1.51e+02 7.12e+01 1.98e+01 0
24 2.53e-01 3.34e+01 1.43e+02 6.96e+01 4.61e+01 0
25 1.26e-01 2.73e+01 1.79e+02 5.00e+01 1.08e+01 0
26 1.26e-01 2.72e+01 1.66e+02 4.82e+01 3.20e+01 0
27 6.31e-02 2.46e+01 1.97e+02 3.71e+01 5.18e+00 0
------------------------- STOP! -------------------------
1 : |fc-fOld| = 6.2891e-01 <= tolF*(1+|f0|) = 1.4241e+02
1 : |xc-x_last| = 5.2038e-01 <= tolX*(1+|x0|) = 3.9461e+00
0 : |proj(x-g)-x| = 5.1758e+00 <= tolG = 1.0000e-01
0 : |proj(x-g)-x| = 5.1758e+00 <= 1e3*eps = 1.0000e-02
0 : maxIter = 100 <= iter = 27
------------------------- DONE! -------------------------
Plot Observed and Predicted Data¶
# Plot the true and apparent resistivities on a sounding curve
fig = plt.figure(figsize=(11, 5))
ax1 = fig.add_axes([0.2, 0.1, 0.6, 0.8])
ax1.semilogy(AB_separations / 2, dobs, "k")
ax1.semilogy(AB_separations / 2, simulation_L2.dpred(recovered_model_L2), "b")
ax1.grid(which="both")
ax1.set_xlabel("AB/2 (m)")
ax1.set_ylabel(r"Apparent Resistivity ($\Omega m$)")
ax1.legend(["Observed", "L2 Inversion"])
plt.show()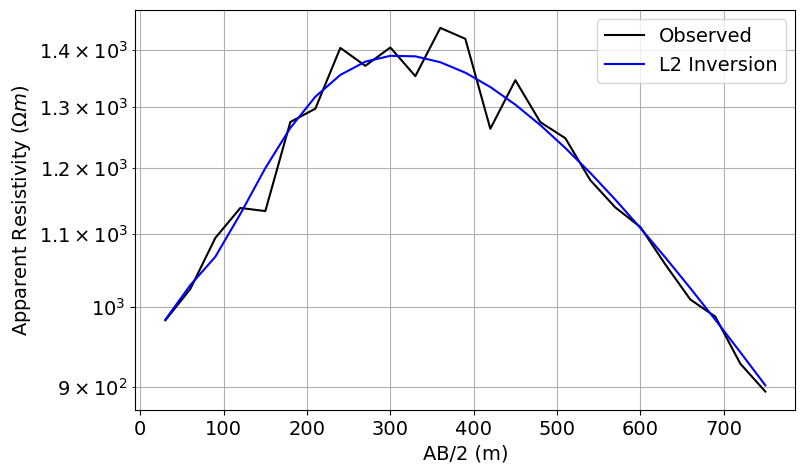
Plot the Recovered Model¶
# Define true model and layer thicknesses
true_resistivities = np.r_[1e3, 4e3, 2e2]
true_layers = np.r_[100.0, 100.0]# Plot true model and recovered model
fig = plt.figure(figsize=(6, 6))
ax1 = fig.add_axes([0.2, 0.15, 0.7, 0.7])
plot_1d_layer_model(true_layers, true_resistivities, ax=ax1, color="k")
plot_1d_layer_model(
layer_thicknesses, log_resistivity_map * recovered_model_L2, ax=ax1, color="b"
)
x_min, x_max = true_resistivities.min(), true_resistivities.max()
ax1.set_xlim(0.9 * x_min, 1.1 * x_max)
ax1.grid()
ax1.set_xlabel(r"Resistivity ($\Omega m$)")
ax1.legend(["True Model", "L2-Model"])
plt.show()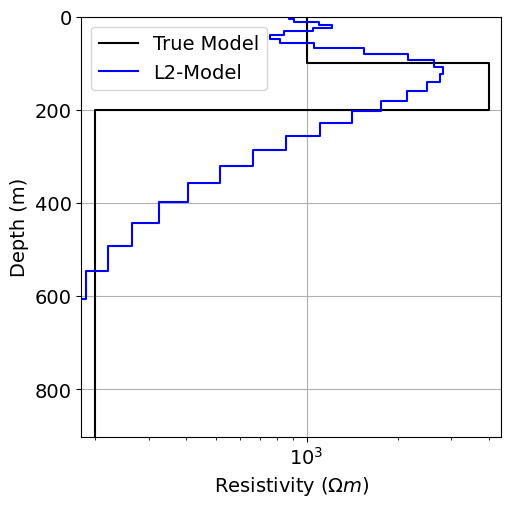
Iteratively Re-weighted Least-Squares Inversion¶
Here, we use the iteratively reweighted least-squares (IRLS) inversion approach to recover sparse and/or blocky models on the set layers.
Define the Forward Simulation¶
simulation_irls = dc.simulation_1d.Simulation1DLayers(
survey=survey,
rhoMap=log_resistivity_map,
thicknesses=layer_thicknesses,
)Define the Data Misfit¶
dmis_irls = data_misfit.L2DataMisfit(simulation=simulation_irls, data=data_object)Define the Regularization¶
Here, we use the Sparse regularization class to constrain the inversion result using an IRLS approach. Here, the scaling constants that balance the smallness and smoothness terms are set directly. Equal emphasis on smallness and smoothness is generally applied by using the inverse square of the smallest cell dimension. The reference model is only applied to the smallness term; which is redundant for the tutorial example since we have set the reference model to an array of zeros. Here, we apply a 1-norm to the smallness term and a 1-norm to first-order smoothness along the x (vertical direction).
reg_irls = regularization.Sparse(
regularization_mesh,
alpha_s=0.1,
alpha_x=1,
reference_model_in_smooth=False,
norms=[1.0, 1.0],
)Define the Optimization Algorithm¶
opt_irls = optimization.InexactGaussNewton(
maxIter=100, maxIterLS=20, cg_maxiter=30, cg_rtol=1e-3
)Define the Inverse Problem¶
inv_prob_irls = inverse_problem.BaseInvProblem(dmis_irls, reg_irls, opt_irls)Provide Inversion Directives¶
Here, we create common directives for IRLS inversion of DC resistivity data and describe their roles. In additon to the UpdateSensitivityWeights, UpdatePreconditioner and BetaEstimate_ByEig directives (described before), inversion with sparse-norms requires the UpdateIRLS directive.
You will notice that we don’t use the BetaSchedule and TargetMisfit directives. Here, the beta cooling schedule is set in the UpdateIRLS directive using the coolingFactor and coolingRate properties. The target misfit for the L2 portion of the IRLS approach is set with the chifact_start property.
sensitivity_weights_irls = directives.UpdateSensitivityWeights(every_iteration=True)
starting_beta_irls = directives.BetaEstimate_ByEig(beta0_ratio=1)
update_jacobi_irls = directives.UpdatePreconditioner(update_every_iteration=True)
update_irls = directives.UpdateIRLS(
cooling_factor=2,
cooling_rate=2,
f_min_change=1e-4,
max_irls_iterations=30,
chifact_start=1.0,
)
directives_list_irls = [
update_irls,
sensitivity_weights_irls,
starting_beta_irls,
update_jacobi_irls,
]Define and Run the Inversion¶
# Here we combine the inverse problem and the set of directives
inv_irls = inversion.BaseInversion(inv_prob_irls, directives_list_irls)
# Run the inversion
recovered_model_irls = inv_irls.run(starting_resistivity_model)INFO: simpeg.InvProblem will set Regularization.reference_model to m0.
INFO: simpeg.InvProblem will set Regularization.reference_model to m0.
Running inversion with SimPEG v0.25.0
============================ Inexact Gauss Newton ============================
# beta phi_d phi_m f |proj(x-g)-x| LS Comment
-----------------------------------------------------------------------------
0 9.07e+02 1.42e+03 0.00e+00 1.42e+03
1 9.07e+02 6.36e+02 2.64e-01 8.75e+02 1.88e+03 0
2 9.07e+02 6.31e+02 2.82e-01 8.87e+02 4.41e+01 0
3 4.54e+02 4.64e+02 5.42e-01 7.10e+02 4.43e+02 0
4 4.54e+02 4.67e+02 5.57e-01 7.20e+02 1.96e+01 0
5 2.27e+02 3.25e+02 1.00e+00 5.52e+02 3.15e+02 0
6 2.27e+02 3.34e+02 1.03e+00 5.66e+02 2.71e+01 0
7 1.13e+02 2.18e+02 1.75e+00 4.16e+02 2.17e+02 0
8 1.13e+02 2.31e+02 1.78e+00 4.33e+02 3.53e+01 0
9 5.67e+01 1.43e+02 2.88e+00 3.06e+02 1.47e+02 0
10 5.67e+01 1.57e+02 2.93e+00 3.23e+02 4.23e+01 0
11 2.83e+01 9.41e+01 4.47e+00 2.21e+02 9.91e+01 0
12 2.83e+01 1.07e+02 4.58e+00 2.37e+02 4.77e+01 0
13 1.42e+01 6.45e+01 6.59e+00 1.58e+02 6.56e+01 0
14 1.42e+01 7.44e+01 6.83e+00 1.71e+02 5.17e+01 0
15 7.09e+00 4.68e+01 9.37e+00 1.13e+02 4.22e+01 0
16 7.09e+00 5.40e+01 9.84e+00 1.24e+02 5.43e+01 0
17 3.54e+00 3.65e+01 1.30e+01 8.24e+01 2.61e+01 0
18 3.54e+00 3.86e+01 1.29e+01 8.45e+01 5.13e+01 0
19 1.77e+00 2.97e+01 1.68e+01 5.95e+01 1.42e+01 0
20 1.77e+00 2.96e+01 1.55e+01 5.71e+01 3.69e+01 0
21 8.86e-01 2.56e+01 1.89e+01 4.23e+01 7.51e+00 0
22 8.86e-01 2.55e+01 1.76e+01 4.11e+01 2.74e+01 0
23 4.43e-01 2.39e+01 2.04e+01 3.30e+01 3.37e+00 0
Reached starting chifact with l2-norm regularization: Start IRLS steps...
irls_threshold 2.266867958619504
24 4.43e-01 2.49e+01 2.42e+01 3.56e+01 2.63e+01 0
25 4.43e-01 2.44e+01 2.46e+01 3.53e+01 3.62e+01 0
26 4.43e-01 2.50e+01 2.82e+01 3.75e+01 4.09e+00 0
27 4.43e-01 2.49e+01 2.83e+01 3.74e+01 1.17e+01 0
28 4.43e-01 2.56e+01 3.23e+01 3.99e+01 2.21e+00 0
29 4.43e-01 2.54e+01 3.24e+01 3.98e+01 9.51e+00 0
30 4.43e-01 2.63e+01 3.71e+01 4.27e+01 2.46e+00 0
31 4.43e-01 2.61e+01 3.72e+01 4.26e+01 9.81e+00 0
32 4.43e-01 2.72e+01 4.29e+01 4.61e+01 2.97e+00 0
33 4.43e-01 2.70e+01 4.30e+01 4.60e+01 9.96e+00 0
34 4.43e-01 2.83e+01 4.95e+01 5.02e+01 3.56e+00 0
35 3.62e-01 2.70e+01 5.27e+01 4.60e+01 7.13e+00 0
36 3.62e-01 2.83e+01 6.02e+01 5.01e+01 6.00e+00 0
37 2.95e-01 2.70e+01 6.43e+01 4.60e+01 6.97e+00 0
38 2.95e-01 2.83e+01 7.32e+01 4.99e+01 5.92e+00 0
39 2.41e-01 2.70e+01 7.81e+01 4.59e+01 5.27e+00 0
40 2.41e-01 2.81e+01 8.77e+01 4.93e+01 5.46e+00 0
41 1.98e-01 2.70e+01 9.33e+01 4.54e+01 3.81e+00 0
42 1.98e-01 2.78e+01 1.02e+02 4.80e+01 4.62e+00 0
43 1.63e-01 2.68e+01 1.08e+02 4.44e+01 3.00e+00 0
44 1.63e-01 2.74e+01 1.16e+02 4.63e+01 3.89e+00 0
45 1.63e-01 2.73e+01 1.15e+02 4.61e+01 3.85e+00 0
46 1.63e-01 2.77e+01 1.22e+02 4.76e+01 1.90e+00 0
47 1.35e-01 2.67e+01 1.27e+02 4.39e+01 2.23e+00 0
48 1.35e-01 2.71e+01 1.34e+02 4.51e+01 3.53e+00 0
49 1.35e-01 2.70e+01 1.33e+02 4.49e+01 2.90e+00 0
50 1.35e-01 2.72e+01 1.37e+02 4.56e+01 1.37e+00 0
51 1.35e-01 2.72e+01 1.35e+02 4.54e+01 2.72e+00 0
52 1.35e-01 2.73e+01 1.38e+02 4.59e+01 1.27e+00 0
53 1.35e-01 2.72e+01 1.37e+02 4.57e+01 2.54e+00 0
54 1.35e-01 2.73e+01 1.38e+02 4.60e+01 1.23e+00 0
55 1.35e-01 2.73e+01 1.37e+02 4.58e+01 2.36e+00 0
56 1.35e-01 2.73e+01 1.39e+02 4.61e+01 1.30e+00 0
57 1.35e-01 2.73e+01 1.38e+02 4.60e+01 2.06e+00 1
58 1.35e-01 2.79e+01 1.55e+02 4.89e+01 4.00e+00 0
59 1.11e-01 2.71e+01 1.63e+02 4.52e+01 2.72e+00 0
60 1.11e-01 2.71e+01 1.61e+02 4.50e+01 3.46e+00 0
61 1.11e-01 2.71e+01 1.60e+02 4.49e+01 1.29e+00 1
62 1.11e-01 2.74e+01 1.71e+02 4.63e+01 2.47e+00 0
63 1.11e-01 2.74e+01 1.71e+02 4.64e+01 4.42e-01 1
64 1.11e-01 2.75e+01 1.75e+02 4.69e+01 1.55e+00 0
65 9.20e-02 2.69e+01 1.82e+02 4.36e+01 2.71e+00 0
66 9.20e-02 2.67e+01 1.77e+02 4.30e+01 2.92e+00 0
67 9.20e-02 2.67e+01 1.75e+02 4.29e+01 1.34e+00 1
68 9.20e-02 2.69e+01 1.86e+02 4.40e+01 2.26e+00 0
69 9.20e-02 2.69e+01 1.86e+02 4.40e+01 1.78e-01 1
70 9.20e-02 2.70e+01 1.88e+02 4.42e+01 1.36e+00 0
71 9.20e-02 2.70e+01 1.88e+02 4.42e+01 2.04e-01 1
72 9.20e-02 2.70e+01 1.90e+02 4.45e+01 1.38e+00 0
73 9.20e-02 2.70e+01 1.90e+02 4.45e+01 1.66e-01 1
74 9.20e-02 2.71e+01 1.91e+02 4.47e+01 1.39e+00 0
75 9.20e-02 2.71e+01 1.91e+02 4.46e+01 1.35e-01 1
76 9.20e-02 2.71e+01 1.92e+02 4.48e+01 1.40e+00 0
77 9.20e-02 2.71e+01 1.92e+02 4.48e+01 1.11e-01 1
78 9.20e-02 2.71e+01 1.93e+02 4.49e+01 1.41e+00 0
------------------------- STOP! -------------------------
1 : |fc-fOld| = 4.4757e-02 <= tolF*(1+|f0|) = 1.4241e+02
1 : |xc-x_last| = 1.7431e+00 <= tolX*(1+|x0|) = 3.9461e+00
1 : |proj(x-g)-x| = 9.2141e-02 <= tolG = 1.0000e-01
0 : |proj(x-g)-x| = 9.2141e-02 <= 1e3*eps = 1.0000e-02
0 : maxIter = 100 <= iter = 78
------------------------- DONE! -------------------------
Plot Observed and Predicted Data¶
# Plot the true and apparent resistivities on a sounding curve
fig = plt.figure(figsize=(11, 5))
ax1 = fig.add_axes([0.2, 0.1, 0.6, 0.8])
ax1.semilogy(AB_separations / 2, dobs, "k")
ax1.semilogy(AB_separations / 2, simulation_L2.dpred(recovered_model_L2), "b")
ax1.semilogy(AB_separations / 2, simulation_irls.dpred(recovered_model_irls), "r")
ax1.grid(which="both")
ax1.set_xlabel("AB/2 (m)")
ax1.set_ylabel(r"Apparent Resistivity ($\Omega m$)")
ax1.legend(["Observed", "L2 Inversion", "IRLS Inversion"])
plt.show()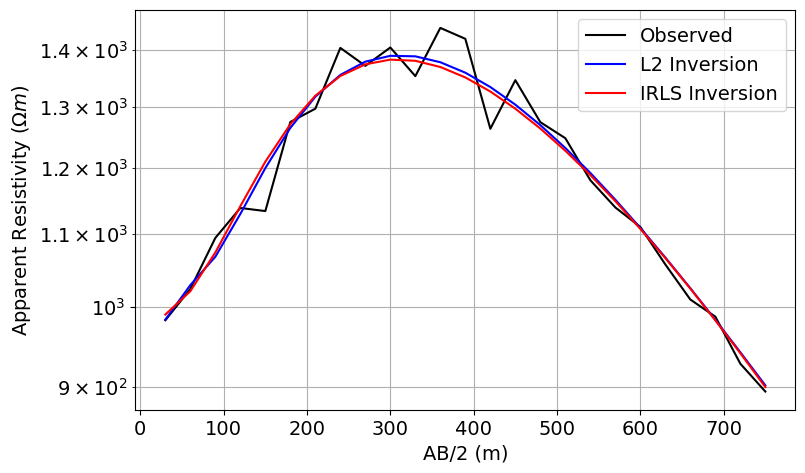
Plot Recovered Models¶
# Plot true model and recovered model
fig = plt.figure(figsize=(6, 6))
ax1 = fig.add_axes([0.2, 0.15, 0.7, 0.7])
plot_1d_layer_model(true_layers, true_resistivities, ax=ax1, color="k")
plot_1d_layer_model(
layer_thicknesses, log_resistivity_map * recovered_model_L2, ax=ax1, color="b"
)
plot_1d_layer_model(
layer_thicknesses, log_resistivity_map * recovered_model_irls, ax=ax1, color="r"
)
x_min, x_max = true_resistivities.min(), true_resistivities.max()
ax1.set_xlim(0.9 * x_min, 1.1 * x_max)
ax1.grid()
ax1.set_xlabel(r"Resistivity ($\Omega m$)")
ax1.legend(["True Model", "L2 Model", "IRLS Model"])
plt.show()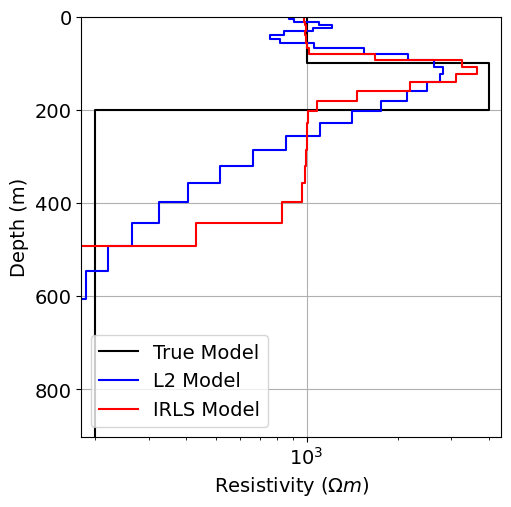
Parametric Inversion¶
Here, we assume the subsurface is defined by a 3-layered Earth. However, the electrical properties and thicknesses of the layers are unknown. Here, we define our model to include log-conductivities and log-thicknesses. When including quantities that span different scales, it is frequently best to define the model in terms of log-values so that each quantity influences the predicted data evenly.
Models and Mappings¶
For a 3-layered Earth model, the model consists of 2 log-thicknesses and 3 log-conductivities. Similar to the 1D Forward Simulation of DC Resistivity Data tutorial, need a mapping that extract log-thicknesses and log-conductivities from the model, and mappings that convert log-values to property values. For this, we require the simpeg.maps.Wires mapping and simpeg.maps.ExpMap mapping classes. Note that successive mappings can be chained together using the operator.
# Wire maps to extract log-thicknesses and log-conductivities
wire_map = maps.Wires(("log_thicknesses", 2), ("log_conductivity", 3))
# Maping for layer thicknesses
log_thicknesses_map = maps.ExpMap() * wire_map.log_thicknesses
# Mapping for conductivities
log_conductivity_map = maps.ExpMap() * wire_map.log_conductivityStarting and Reference Models¶
This problem is highly non-linear so it is important to have a reasonable estimate of the true model.
starting_parametric_model = np.log(np.r_[125.0, 50.0, 1e-3, 2e-3, 5e-2])
reference_parametric_model = starting_parametric_model.copy()Define the Forward Simulation¶
Because the layer thicknesses are part of the model, we define the thicknessesMap. Because we are working in terms of electrical conductivity, we must define the sigmaMap.
simulation_parametric = dc.simulation_1d.Simulation1DLayers(
survey=survey,
sigmaMap=log_conductivity_map,
thicknessesMap=log_thicknesses_map,
)Define the Data Misfit¶
dmis_parametric = data_misfit.L2DataMisfit(
simulation=simulation_parametric, data=data_object
)Define a (Combo) Regularization¶
We need to define a regularization for each model parameter type. In this case, we have log-thicknesses and log-conductivities. For each model parameter type, we create a 1D tensor mesh with length equal to the number of parameters. In the mapping keyword argument, we used the wire map that extracts the specific model parameters from the model.
Using the operator, separate regularizations can be summed to form a regularization that is also a ComboObjectiveFunction. By setting the multipliers property, we can emphasis the relative contributions of the log-thicknesses and log-conductivities regularizations.
reg_1 = regularization.Smallness(
TensorMesh([(np.ones(2))], "0"),
mapping=wire_map.log_thicknesses,
reference_model=reference_parametric_model,
)
reg_2 = regularization.Smallness(
TensorMesh([(np.ones(3))], "0"),
mapping=wire_map.log_conductivity,
reference_model=reference_parametric_model,
)
reg_parametric = reg_1 + reg_2
reg_parametric.multipliers = [1.0, 0.1]Define the Optimization Algorithm¶
opt_parametric = optimization.InexactGaussNewton(
maxIter=100, maxIterLS=20, cg_maxiter=20, cg_rtol=1e-3
)Define the Inverse Problem¶
inv_prob_parametric = inverse_problem.BaseInvProblem(
dmis_parametric, reg_parametric, opt_parametric
)Provide Inversion Directives¶
sensitivity_weights = directives.UpdateSensitivityWeights(every_iteration=True)
update_jacobi = directives.UpdatePreconditioner(update_every_iteration=True)
starting_beta = directives.BetaEstimate_ByEig(beta0_ratio=10)
beta_schedule = directives.BetaSchedule(coolingFactor=2.0, coolingRate=2)
target_misfit = directives.TargetMisfit(chifact=1.0)
directives_list_parametric = [
sensitivity_weights,
update_jacobi,
starting_beta,
beta_schedule,
target_misfit,
]Define and Run Inversion¶
inv_parametric = inversion.BaseInversion(
inv_prob_parametric, directives_list_parametric
)
recovered_model_parametric = inv_parametric.run(starting_parametric_model)INFO: Directive TargetMisfit: Target data misfit is 25.0
Running inversion with SimPEG v0.25.0
============================ Inexact Gauss Newton ============================
# beta phi_d phi_m f |proj(x-g)-x| LS Comment
-----------------------------------------------------------------------------
0 1.35e+05 1.95e+04 0.00e+00 1.95e+04
1 1.35e+05 1.34e+04 1.48e-02 1.54e+04 1.72e+04 0
2 1.35e+05 1.23e+04 2.80e-02 1.61e+04 3.55e+03 0
3 6.76e+04 7.08e+03 7.50e-02 1.22e+04 8.79e+03 0
4 6.76e+04 6.78e+03 1.05e-01 1.39e+04 1.24e+03 0
5 3.38e+04 2.53e+03 1.83e-01 8.70e+03 7.27e+03 0
6 3.38e+04 2.72e+03 2.22e-01 1.02e+04 1.31e+03 0
7 1.69e+04 8.76e+02 2.89e-01 5.77e+03 4.93e+03 0
8 1.69e+04 9.73e+02 3.17e-01 6.32e+03 9.58e+02 0
9 8.45e+03 3.38e+02 3.76e-01 3.52e+03 2.93e+03 0
10 8.45e+03 3.52e+02 4.03e-01 3.75e+03 5.28e+02 0
11 4.22e+03 1.36e+02 4.63e-01 2.09e+03 1.70e+03 0
12 4.22e+03 1.37e+02 4.99e-01 2.24e+03 2.63e+02 0
13 2.11e+03 6.22e+01 5.41e-01 1.21e+03 9.90e+02 0
14 2.11e+03 6.29e+01 5.76e-01 1.28e+03 1.33e+02 0
15 1.06e+03 3.66e+01 5.99e-01 6.69e+02 5.60e+02 0
16 1.06e+03 3.70e+01 6.29e-01 7.01e+02 6.69e+01 0
17 5.28e+02 2.78e+01 6.41e-01 3.67e+02 3.05e+02 0
18 5.28e+02 2.80e+01 6.64e-01 3.79e+02 2.66e+01 0
19 2.64e+02 2.49e+01 6.72e-01 2.02e+02 1.61e+02 0
------------------------- STOP! -------------------------
1 : |fc-fOld| = 1.0093e+00 <= tolF*(1+|f0|) = 1.9497e+03
1 : |xc-x_last| = 4.6359e-02 <= tolX*(1+|x0|) = 1.2573e+00
0 : |proj(x-g)-x| = 1.6138e+02 <= tolG = 1.0000e-01
0 : |proj(x-g)-x| = 1.6138e+02 <= 1e3*eps = 1.0000e-02
0 : maxIter = 100 <= iter = 19
------------------------- DONE! -------------------------
Plot Observed and Predicted Data¶
# Plot the true and apparent resistivities on a sounding curve
fig = plt.figure(figsize=(11, 5))
ax1 = fig.add_axes([0.2, 0.1, 0.6, 0.8])
ax1.semilogy(AB_separations / 2, dobs, "k")
ax1.semilogy(AB_separations / 2, simulation_L2.dpred(recovered_model_L2), "b")
ax1.semilogy(AB_separations / 2, simulation_irls.dpred(recovered_model_irls), "r")
ax1.semilogy(
AB_separations / 2, simulation_parametric.dpred(recovered_model_parametric), "g"
)
ax1.grid(which="both")
ax1.set_xlabel("AB/2 (m)")
ax1.set_ylabel(r"Apparent Resistivity ($\Omega m$)")
ax1.legend(["Observed", "L2 Inversion", "IRLS Inversion", "Parametric Inversion"])
plt.show()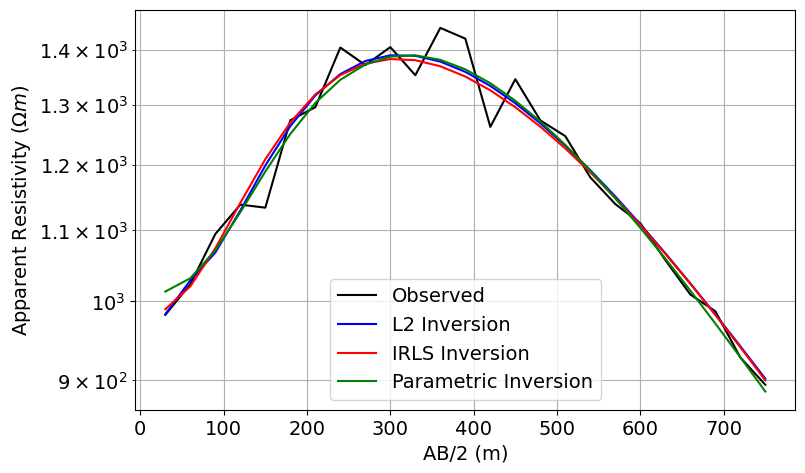
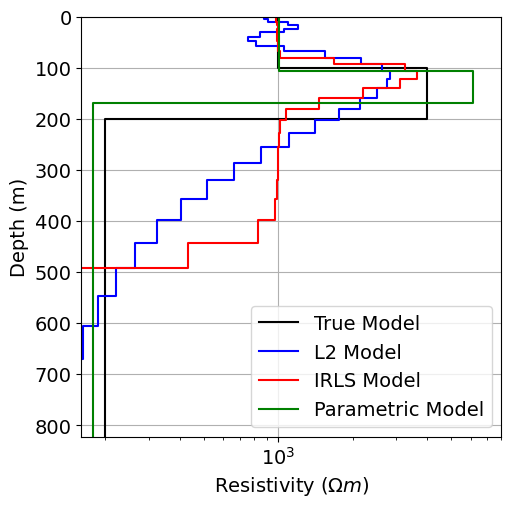
Plot Recovered Models¶
fig = plt.figure(figsize=(6, 6))
ax1 = fig.add_axes([0.2, 0.15, 0.7, 0.7])
plot_1d_layer_model(true_layers, true_resistivities, ax=ax1, color="k")
plot_1d_layer_model(
layer_thicknesses, log_resistivity_map * recovered_model_L2, ax=ax1, color="b"
)
plot_1d_layer_model(
layer_thicknesses, log_resistivity_map * recovered_model_irls, ax=ax1, color="r"
)
plot_1d_layer_model(
log_thicknesses_map * recovered_model_parametric,
1 / (log_conductivity_map * recovered_model_parametric),
ax=ax1,
color="g",
)
x_min, x_max = true_resistivities.min(), true_resistivities.max()
ax1.set_xlim(0.8 * x_min, 2 * x_max)
ax1.set_ylim([np.sum(layer_thicknesses), 0])
ax1.grid()
ax1.set_xlabel(r"Resistivity ($\Omega m$)")
ax1.legend(["True Model", "L2 Model", "IRLS Model", "Parametric Model"])
plt.show()