DC Simulation: Blocks Model#
Geoscientific Problem#
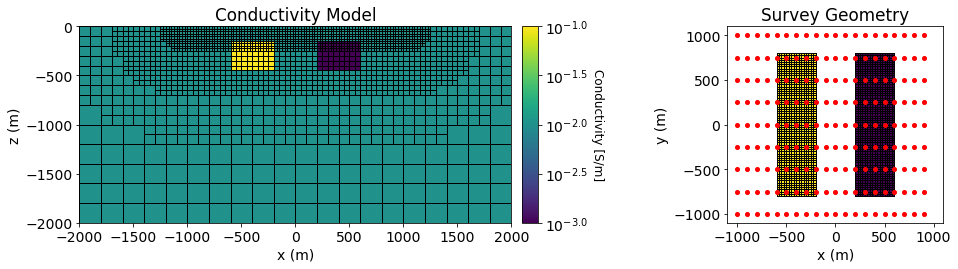
Pole-dipole DC resistivity data were simulated over both a conductive and a resistive block. The background conductivity was \(\sigma_0\) = 0.01 S/m. The conductor had a conductivity of \(\sigma_c\) = 0.1 S/m and the resistor had a conductivity of \(\sigma_r\) = 0.001 S/m. Both blocks were oriented along the Northing direction and had x, y and z dimensions of 400 m, 1600 m and 320 m. Both blocks were buried at a depth of 160 m.
DC voltage data were simulated with a pole-dipole configuration. The survey consisted of 9 West-East survey lines, each with a length of 2000 m. The line spacing was 250 m and the electrode spacing was 100 m.
A figure illustrating the density contrast model and survey geometry is shown further down
Codes/Formulations Being Compared#
SimPEG 3D Nodal Formulation: This approach to solving the forward problem uses the SimPEG.electromagnetics.static.simulation.Simulation3DNodal simulation class.
UBC-GIF DCIP OcTree: DCIP OcTree is a voxel cell DC/IP forward modeling and inversion package developed by the UBC Geophysical Inversion Facility. This software is proprietary and can ONLY be acquired through appropriate commerical licenses. The numerical approach of the forward simulation is described in the online manual’s theory section. If you have a valid license, there are instructions for reproducing the results (add link).
Loading Assets Into the SimPEG Framework#
We start by importing any necessary packages for running the notebook.
Show code cell source
from SimPEG.electromagnetics.static import resistivity as dc
from SimPEG.electromagnetics.static.utils.static_utils import (
plot_3d_pseudosection, apparent_resistivity
)
from SimPEG.utils.io_utils import read_dcipoctree_ubc, write_dcipoctree_ubc
from discretize import TreeMesh
import matplotlib as mpl
import matplotlib.pyplot as plt
import plotly
import numpy as np
mpl.rcParams.update({"font.size": 14})
Next we download the mesh, model and simulated data for each code/formulation.
Show code cell source
# For each package, download .tar files
The mesh, model and predicted data for each code are then loaded into the SimPEG framework for plotting.
Show code cell source
rootdir = './../../../assets/dcip/block_model_dc_fwd_simpeg_octree/'
mesh_simpeg = TreeMesh.readUBC(rootdir+'octree_mesh.txt')
model_simpeg = TreeMesh.readModelUBC(mesh_simpeg, rootdir+'true_model.con')
data_simpeg = read_dcipoctree_ubc(rootdir+'dpred_simpeg.txt', 'volt')
rootdir = './../../../assets/dcip/block_model_dc_fwd_dcipoctree/'
mesh_ubc = TreeMesh.readUBC(rootdir+'octree_mesh.txt')
model_ubc = TreeMesh.readModelUBC(mesh_simpeg, rootdir+'true_model.con')
data_ubc = read_dcipoctree_ubc(rootdir+'data_dc.txt', 'volt')
Plot Geophysical Scenario#
Below, we plot the conductivity model and survey geometry for the forward simulation.
Show code cell source
vmin = np.log10(model_simpeg.min())
vmax = np.log10(model_simpeg.max())
ind = int(len(mesh_simpeg.hy)/2)
fig = plt.figure(figsize=(15, 3.5))
ax1 = fig.add_axes([0.1, 0.12, 0.4, 0.78])
mesh_simpeg.plot_slice(
np.log10(model_simpeg), ax=ax1, normal='Y', grid=True,
ind=ind, clim=(vmin, vmax), pcolorOpts={"cmap": "viridis"},
)
ax1.set_title("Conductivity Model")
ax1.set_xlim([-2000, 2000])
ax1.set_ylim([-2000, 0])
ax1.set_xlabel("x (m)")
ax1.set_ylabel("z (m)")
ax2 = fig.add_axes([0.51, 0.12, 0.015, 0.78])
norm = mpl.colors.Normalize(
vmin=vmin, vmax=vmax
)
cbar = mpl.colorbar.ColorbarBase(
ax2, norm=norm, cmap=mpl.cm.viridis, orientation="vertical", format="$10^{%.1f}$"
)
cbar.set_label("Conductivity [S/m]", rotation=270, labelpad=15, size=12)
ax3 = fig.add_axes([0.7, 0.12, 0.2, 0.78])
ind = int(len(mesh_simpeg.hz)-10)
masked_model = np.log10(model_simpeg)
masked_model[masked_model==-2]=np.NaN
mesh_simpeg.plot_slice(
masked_model, ax=ax3, normal='Z', grid=True,
ind=ind, clim=(vmin, vmax), pcolorOpts={"cmap": "viridis"},
)
for ii in range(0, 9):
ax3.plot(np.arange(-1000, 1000, 100), -1000+ii*250*np.ones(20), 'ro', markersize=4)
ax3.set_xlim([-1100, 1100])
ax3.set_ylim([-1100, 1100])
ax3.set_xlabel('x (m)')
ax3.set_ylabel('y (m)')
ax3.set_title('Survey Geometry')
Text(0.5, 1.0, 'Survey Geometry')
Comparing Simulated Voltage Data#
Here we plot the simulated data for all codes.
Show code cell source
mpl.rcParams.update({'font.size': 16})
plane_points = []
for x in np.arange(-1000, 1100, 500):
p1, p2, p3 = np.array([-1000.,x,0]), np.array([1000,x,0]), np.array([1000,x,-1000])
plane_points.append([p1,p2,p3])
scene = dict(
xaxis=dict(range=[-1000, 1000]),
yaxis=dict(range=[-1000, 1000]),
zaxis=dict(range=[-500, 0]),
aspectratio=dict(x=1, y=1, z=0.5)
)
scene_camera=dict(
center=dict(x=-0.1, y=0, z=-0.2), eye=dict(x=1.2, y=-1, z=1.5)
)
vlim = [data_ubc.dobs.min(), data_ubc.dobs.max()]
fig1 = plot_3d_pseudosection(
data_ubc.survey, data_ubc.dobs, scale='log', vlim=vlim,
plane_points=plane_points, plane_distance=10., units='V'
)
fig1.update_layout(
title_text="Voltage Data (UBC)", title_x=0.5,
scene_camera=scene_camera, scene=scene, width=600, height=550
)
plotly.io.show(fig1)
vlim = [data_simpeg.dobs.min(), data_simpeg.dobs.max()]
fig2 = plot_3d_pseudosection(
data_simpeg.survey, data_simpeg.dobs, marker_size=150, scale='log', vlim=vlim,
plane_points=plane_points, plane_distance=10., units='V'
)
fig2.update_layout(
title_text="Voltage Data (SimPEG)", title_x=0.5,
scene_camera=scene_camera, scene=scene, width=600, height=550
)
plotly.io.show(fig2)
Comparing Apparent Conductivities#
To better illustrate the simulated data, we plot in terms of apparent conductivities.
Show code cell source
mpl.rcParams.update({'font.size': 16})
plane_points = []
for x in np.arange(-1000, 1100, 500):
p1, p2, p3 = np.array([-1000.,x,0]), np.array([1000,x,0]), np.array([1000,x,-1000])
plane_points.append([p1,p2,p3])
scene_camera=dict(
center=dict(x=-0.1, y=0, z=-0.2), eye=dict(x=1.2, y=-1, z=1.5)
)
scene = dict(
xaxis=dict(range=[-1000, 1000]),
yaxis=dict(range=[-1000, 1000]),
zaxis=dict(range=[-500, 0]),
aspectratio=dict(x=1, y=1, z=0.5)
)
app_cond_ubc = 1./apparent_resistivity(
data_ubc, space_type="half space", dobs=data_ubc.dobs, eps=1e-10,
)
vlim = [app_cond_ubc.min(), app_cond_ubc.max()]
fig1 = plot_3d_pseudosection(
data_ubc.survey, app_cond_ubc, scale='log', vlim=vlim,
plane_points=plane_points, plane_distance=10., units='S/m'
)
fig1.update_layout(
title_text="Apparent Conductivities (UBC)", title_x=0.5,
width=600, height=550, scene_camera=scene_camera, scene=scene
)
plotly.io.show(fig1)
app_cond_simpeg = 1./apparent_resistivity(
data_simpeg, space_type="half space", dobs=data_simpeg.dobs, eps=1e-10,
)
vlim = [app_cond_simpeg.min(), app_cond_simpeg.max()]
fig2 = plot_3d_pseudosection(
data_simpeg.survey, app_cond_simpeg, scale='log', vlim=vlim,
plane_points=plane_points, plane_distance=10., units='S/m'
)
fig2.update_layout(
title_text="Apparent Conductivities (SimPEG)", title_x=0.5,
width=600, height=550, scene_camera=scene_camera, scene=scene
)
plotly.io.show(fig2)
Percent Error Plot#
Show code cell source
err = 100*(data_simpeg.dobs - data_ubc.dobs)/data_simpeg.dobs
vlim = [err.min(), err.max()]
fig = plot_3d_pseudosection(
data_simpeg.survey, err, scale='linear', vlim=vlim,
plane_points=plane_points, plane_distance=10., units='%'
)
fig.update_layout(
title_text="% Error", title_x=0.5, width=600, height=550, scene_camera=scene_camera, scene=scene
)
plotly.io.show(fig)
print('Min % error:', np.min(np.abs(err)))
print('Max % error:', np.max(np.abs(err)))
print('Average % error:', np.mean(np.abs(err)))
print('Median % error:', np.median(np.abs(err)))
Min % error: 0.005245189395426906
Max % error: 6.154689069491367
Average % error: 2.20696037763589
Median % error: 2.265438988175177
