Reproduce: SimPEG#
Inverting Gravity Data Over a Block in a Halfspace: Compact and Blocky Model#
Here, we invert gravity anomaly data collected over a block within a homogeneous halfspace. We invert for a compact and blocky model using an iteratively re-weighted least-squares inversion approach.
The true model consists of a denser block (0.1 \(g/cm^3\)) within a halfspace (0 \(g/cm^3\)). The dimensions of the block in the x, y and z directions are all 200 m. The block is buried at a depth of 200 m.
The data being inverted were generated using the UBC-GIF GRAV3D v6.0.1 code. Synthetic gravity data were simulated at a heigh 1 m above the surface within a 1000 m by 1000 m region; the center of which lied directly over the center of the block. Gaussian noise with a standard deviation of 0.002 mGal were added to the synthetic data. Uncertainties of 0.002 mGal were assigned to the data before inverting.
Background Theory#
Link to the docstrings for the simulation class. The docstrings will have a citation and show the integral equation.
Reproducing the Inversion Result#
We begin by importing all necessary Python packages for running the notebook.
Show code cell source
from SimPEG import dask
from SimPEG.potential_fields import gravity
from SimPEG.utils import plot2Ddata
from SimPEG.utils.io_utils import read_grav3d_ubc, write_grav3d_ubc
from SimPEG import maps, data, data_misfit, regularization, optimization, inverse_problem, inversion, directives
from discretize import TensorMesh
import matplotlib as mpl
import matplotlib.pyplot as plt
import numpy as np
mpl.rcParams.update({"font.size": 14})
write_output = True
A compressed folder containing the assets required to run the notebook is then downloaded. This includes the mesh, true model, and observed data files.
Show code cell source
# Download .tar file
Extracted files are then loaded into the SimPEG framework.
Show code cell source
rootdir = './../../../assets/gravity/block_halfspace_gravity_inv_sparse_simpeg/'
meshfile = rootdir + 'mesh.txt'
truemodelfile = rootdir + 'true_model.den'
obsfile = rootdir + 'dobs.grv'
sensitivitydir = './block_halfspace_gravity_inv_sparse_simpeg/'
mesh = TensorMesh.read_UBC(meshfile)
true_model = TensorMesh.read_model_UBC(mesh, truemodelfile)
grav_data = read_grav3d_ubc(obsfile)
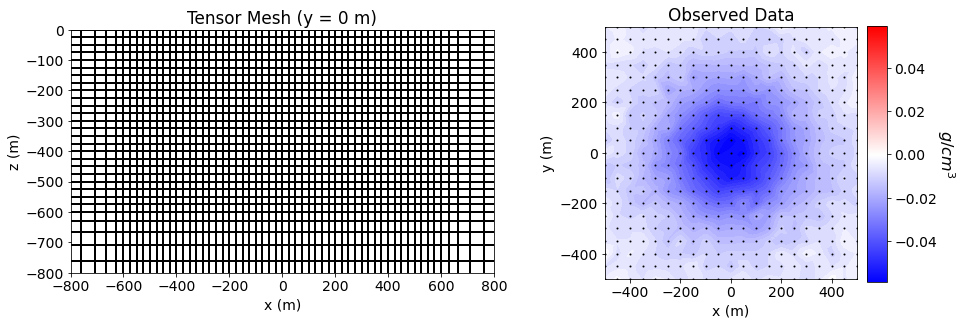
We then plot the observed data and the mesh on which we will recover a density contrast model.
Show code cell source
fig = plt.figure(figsize=(14, 4.5))
ax11 = fig.add_axes([0.1, 0.15, 0.42, 0.75])
ind = int(mesh.shape_cells[1]/2)
nan_array = np.zeros(len(true_model))
nan_array[:] = np.NaN
mesh.plot_slice(
nan_array, normal='Y', ind=ind, ax=ax11,
pcolor_opts={"cmap": mpl.cm.binary}, grid=True,
)
ax11.set_xlim([-800, 800])
ax11.set_ylim([-800, 0])
ax11.set_title("Tensor Mesh (y = 0 m)")
ax11.set_xlabel("x (m)")
ax11.set_ylabel("z (m)")
ax21 = fig.add_axes([0.63, 0.12, 0.25, 0.8])
xyz = grav_data.survey.receiver_locations
max_val = np.max(np.abs(grav_data.dobs))
plot2Ddata(
xyz, grav_data.dobs, ax=ax21, dataloc=True, ncontour=50,
clim=(-max_val, max_val), contourOpts={"cmap": "bwr"}
)
ax21.set_title("Observed Data")
ax21.set_xlabel("x (m)")
ax21.set_ylabel("y (m)")
ax22 = fig.add_axes([0.89, 0.12, 0.02, 0.79])
norm = mpl.colors.Normalize(vmin=-max_val, vmax=max_val)
cbar = mpl.colorbar.ColorbarBase(
ax22, norm=norm, orientation="vertical", cmap=mpl.cm.bwr
)
cbar.set_label("$g/cm^3$", rotation=270, labelpad=20, size=16)
plt.show()
<__array_function__ internals>:5: UserWarning: Warning: converting a masked element to nan.
C:\Users\devin\anaconda3\lib\site-packages\numpy\core\_asarray.py:102: UserWarning: Warning: converting a masked element to nan.
return array(a, dtype, copy=False, order=order)
Next, we define the mapping from the model space to the mesh and the simulation.
rho_map = maps.IdentityMap(nP=mesh.nC)
simulation = gravity.simulation.Simulation3DIntegral(
survey=grav_data.survey,
mesh=mesh,
rhoMap=rho_map,
store_sensitivities="disk",
sensitivity_path=sensitivitydir
)
We now define a starting model and reference model for the inversion.
# Starting and reference model
mref = np.zeros(mesh.nC)
m0 = 1e-4*np.ones(mesh.nC)
Here we define the measure of data misfit, the regularization and the algorithm used to compute the step-direction at each iteration. These are used to define the inverse problem.
Show code cell source
dmis = data_misfit.L2DataMisfit(data=grav_data, simulation=simulation)
reg_map = maps.IdentityMap(nP=mesh.nC)
reg = regularization.Sparse(
mesh, mapping=reg_map, reference_model=mref, gradient_type='components',
alpha_s=1e-3, alpha_x=1, alpha_y=1, alpha_z=1
)
reg.norms = np.r_[0., 1., 1., 1.]
opt = optimization.ProjectedGNCG(
maxIter=50, maxIterCG=50, tolCG=1e-4
)
inv_prob = inverse_problem.BaseInvProblem(dmis, reg, opt)
Here, we define the directives for the inversion
Show code cell source
starting_beta = directives.BetaEstimate_ByEig(beta0_ratio=200.)
beta_schedule = directives.BetaSchedule(coolingFactor=2, coolingRate=1)
save_iteration = directives.SaveOutputEveryIteration(save_txt=False)
update_IRLS = directives.Update_IRLS(
max_irls_iterations=35, chifact_start=1., f_min_change=1e-3
)
update_jacobi = directives.UpdatePreconditioner()
sensitivity_weights = directives.UpdateSensitivityWeights(everyIter=True)
directives_list = [
sensitivity_weights,
starting_beta,
beta_schedule,
save_iteration,
update_IRLS,
update_jacobi,
]
Finally, we define and run the inversion.
inv = inversion.BaseInversion(inv_prob, directives_list)
simpeg_model = inv.run(m0)
simpeg_model = rho_map*simpeg_model
dpred = inv_prob.dpred
SimPEG.InvProblem is setting bfgsH0 to the inverse of the eval2Deriv.
***Done using the default solver Pardiso and no solver_opts.***
model has any nan: 0
=============================== Projected GNCG ===============================
# beta phi_d phi_m f |proj(x-g)-x| LS Comment
-----------------------------------------------------------------------------
x0 has any nan: 0
0 1.28e+06 1.80e+04 7.09e-04 1.89e+04 1.40e+05 0
1 3.20e+05 1.12e+04 3.31e-03 1.23e+04 8.63e+04 0
2 7.99e+04 6.33e+03 1.11e-02 7.22e+03 4.67e+04 0 Skip BFGS
3 2.00e+04 2.79e+03 3.44e-02 3.48e+03 2.52e+04 0 Skip BFGS
4 5.00e+03 7.62e+02 8.32e-02 1.18e+03 1.11e+04 0 Skip BFGS
5 1.25e+03 2.33e+02 1.31e-01 3.96e+02 4.14e+03 0 Skip BFGS
Reached starting chifact with l2-norm regularization: Start IRLS steps...
irls_threshold 0.009973604628269106
6 3.12e+02 1.10e+02 3.01e-01 2.05e+02 1.60e+03 0 Skip BFGS
7 4.78e+02 5.35e+01 5.55e-01 3.19e+02 1.68e+03 0 Skip BFGS
8 5.27e+02 9.14e+01 5.68e-01 3.91e+02 1.16e+03 0
9 5.18e+02 1.14e+02 6.66e-01 4.59e+02 1.31e+03 0
10 4.68e+02 1.37e+02 7.72e-01 4.98e+02 1.36e+03 0
11 3.98e+02 1.57e+02 8.32e-01 4.89e+02 9.66e+02 0
12 3.29e+02 1.69e+02 8.70e-01 4.55e+02 6.14e+02 0 Skip BFGS
13 2.69e+02 1.74e+02 9.03e-01 4.17e+02 4.01e+02 0 Skip BFGS
14 2.18e+02 1.76e+02 9.22e-01 3.77e+02 2.33e+02 0 Skip BFGS
15 1.78e+02 1.75e+02 9.40e-01 3.42e+02 1.56e+02 0 Skip BFGS
16 1.46e+02 1.73e+02 9.58e-01 3.13e+02 1.71e+02 0 Skip BFGS
17 1.20e+02 1.70e+02 1.00e+00 2.90e+02 2.08e+02 0 Skip BFGS
18 9.94e+01 1.67e+02 1.03e+00 2.69e+02 2.75e+02 0 Skip BFGS
19 8.32e+01 1.64e+02 1.05e+00 2.51e+02 3.26e+02 0 Skip BFGS
20 7.04e+01 1.59e+02 1.07e+00 2.34e+02 3.94e+02 0 Skip BFGS
21 6.06e+01 1.53e+02 1.12e+00 2.21e+02 4.45e+02 0 Skip BFGS
22 5.31e+01 1.46e+02 1.18e+00 2.09e+02 4.95e+02 0 Skip BFGS
23 4.79e+01 1.37e+02 1.23e+00 1.96e+02 5.21e+02 0 Skip BFGS
24 4.47e+01 1.27e+02 1.27e+00 1.84e+02 5.11e+02 0 Skip BFGS
25 4.33e+01 1.17e+02 1.29e+00 1.73e+02 4.78e+02 0 Skip BFGS
26 4.38e+01 1.08e+02 1.33e+00 1.66e+02 4.13e+02 0 Skip BFGS
27 4.57e+01 1.01e+02 1.34e+00 1.63e+02 3.73e+02 0 Skip BFGS
28 4.87e+01 9.74e+01 1.33e+00 1.62e+02 3.68e+02 0 Skip BFGS
29 5.24e+01 9.55e+01 1.28e+00 1.63e+02 3.88e+02 0 Skip BFGS
30 5.65e+01 9.53e+01 1.23e+00 1.65e+02 4.15e+02 0
31 6.07e+01 9.61e+01 1.16e+00 1.66e+02 4.37e+02 0
32 6.48e+01 9.72e+01 1.11e+00 1.69e+02 4.63e+02 0
33 6.85e+01 9.90e+01 1.07e+00 1.72e+02 4.91e+02 0
34 7.14e+01 1.01e+02 1.04e+00 1.75e+02 4.95e+02 0
35 7.35e+01 1.04e+02 1.00e+00 1.78e+02 4.82e+02 0
36 7.48e+01 1.06e+02 9.77e-01 1.80e+02 4.64e+02 0
37 7.54e+01 1.08e+02 9.56e-01 1.81e+02 4.41e+02 0 Skip BFGS
38 7.55e+01 1.10e+02 9.40e-01 1.81e+02 4.15e+02 0 Skip BFGS
39 7.53e+01 1.11e+02 9.28e-01 1.81e+02 3.93e+02 0 Skip BFGS
40 7.49e+01 1.11e+02 9.17e-01 1.80e+02 3.79e+02 0 Skip BFGS
Reach maximum number of IRLS cycles: 35
------------------------- STOP! -------------------------
1 : |fc-fOld| = 0.0000e+00 <= tolF*(1+|f0|) = 1.8905e+03
1 : |xc-x_last| = 2.5615e-02 <= tolX*(1+|x0|) = 1.0397e-01
0 : |proj(x-g)-x| = 3.7930e+02 <= tolG = 1.0000e-01
0 : |proj(x-g)-x| = 3.7930e+02 <= 1e3*eps = 1.0000e-02
0 : maxIter = 50 <= iter = 41
------------------------- DONE! -------------------------
If desired, we can output the recovered model and the predicted data.
if write_output:
TensorMesh.write_model_UBC(mesh, rootdir+'recovered_model.den', simpeg_model)
data_dpred = data.Data(survey=grav_data.survey, dobs=dpred)
write_grav3d_ubc(rootdir+'dpred.grv', data_dpred)
Observation file saved to: ./../../../assets/gravity/block_halfspace_gravity_inv_sparse_simpeg/dpred.grv
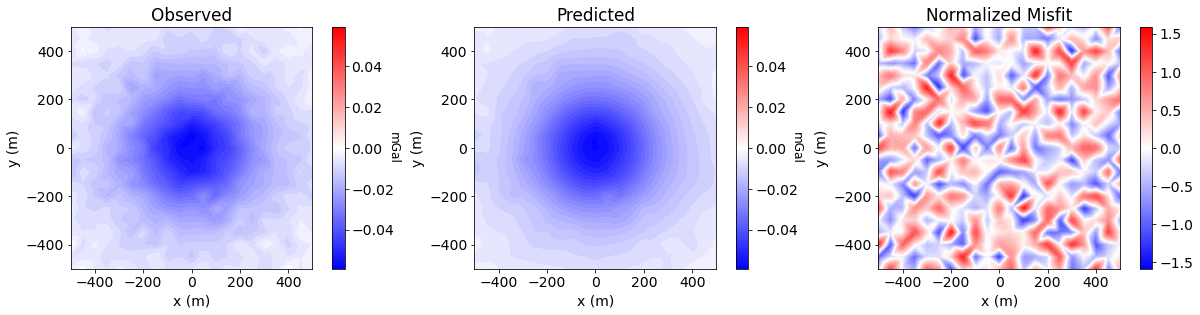
Data Misfit#
Show code cell source
data_array = np.c_[grav_data.dobs, dpred, (grav_data.dobs-dpred)/grav_data.standard_deviation]
fig = plt.figure(figsize=(17, 4))
plot_title = ["Observed", "Predicted", "Normalized Misfit"]
plot_units = ["mGal", "mGal", ""]
ax1 = 3 * [None]
ax2 = 3 * [None]
norm = 3 * [None]
cbar = 3 * [None]
cplot = 3 * [None]
v_lim = [
np.max(np.abs(grav_data.dobs)),
np.max(np.abs(grav_data.dobs)),
np.max(np.abs(data_array[:, 2]))
]
for ii in range(0, 3):
ax1[ii] = fig.add_axes([0.33 * ii + 0.03, 0.11, 0.25, 0.84])
cplot[ii] = plot2Ddata(
xyz,
data_array[:, ii],
ax=ax1[ii],
ncontour=50,
clim=(-v_lim[ii], v_lim[ii]),
contourOpts={"cmap": "bwr"}
)
ax1[ii].set_title(plot_title[ii])
ax1[ii].set_xlabel("x (m)")
ax1[ii].set_ylabel("y (m)")
ax2[ii] = fig.add_axes([0.33 * ii + 0.27, 0.11, 0.01, 0.84])
norm[ii] = mpl.colors.Normalize(vmin=-v_lim[ii], vmax=v_lim[ii])
cbar[ii] = mpl.colorbar.ColorbarBase(
ax2[ii], norm=norm[ii], orientation="vertical", cmap=mpl.cm.bwr
)
cbar[ii].set_label(plot_units[ii], rotation=270, labelpad=5, size=12)
plt.show()
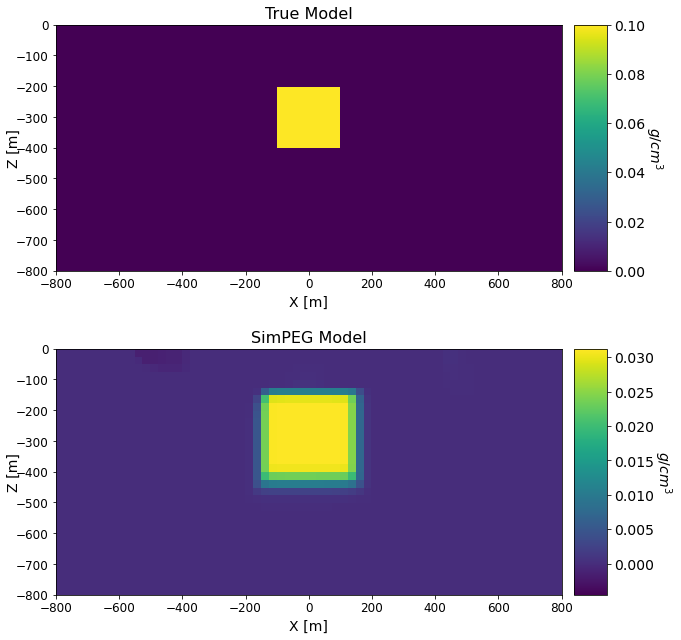
Comparing True and Recovered Models#
Show code cell source
fig = plt.figure(figsize=(9, 9))
font_size = 14
models_list = [true_model, simpeg_model]
titles_list = ['True Model', 'SimPEG Model']
ax1 = 2*[None]
cplot = 2*[None]
ax2 = 2*[None]
cbar = 2*[None]
for qq in range(0, 2):
ax1[qq] = fig.add_axes([0.1, 0.55 - 0.5*qq, 0.78, 0.38])
cplot[qq] = mesh.plot_slice(
models_list[qq], normal='Y', ind=int(mesh.shape_cells[1]/2), grid=False, ax=ax1[qq]
)
cplot[qq][0].set_clim((np.min(models_list[qq]), np.max(models_list[qq])))
ax1[qq].set_xlim([-800, 800])
ax1[qq].set_ylim([-800, 0])
ax1[qq].set_xlabel("X [m]", fontsize=font_size)
ax1[qq].set_ylabel("Z [m]", fontsize=font_size, labelpad=-5)
ax1[qq].tick_params(labelsize=font_size - 2)
ax1[qq].set_title(titles_list[qq], fontsize=font_size + 2)
ax2[qq] = fig.add_axes([0.9, 0.55 - 0.5*qq, 0.05, 0.38])
norm = mpl.colors.Normalize(vmin=np.min(models_list[qq]), vmax=np.max(models_list[qq]))
cbar[qq] = mpl.colorbar.ColorbarBase(
ax2[qq], norm=norm, orientation="vertical"
)
cbar[qq].set_label(
"$g/cm^3$",
rotation=270,
labelpad=20,
size=font_size,
)
plt.show()