Reproduce: SimPEG OcTree#
Inverting Pole-Dipole IP Data over a Conductive (and Chargeable) and a Resistive Block#
Here, we invert pole-dipole IP data collected over a conductive and a resistive block. The conductive block is also chargeable. We invert for an intrinsic chargeability model using a least-squares inversion approach.
For the true model, the background conductivity \(\sigma_0\) = 0.01 S/m. The conductor has a conductivity of \(\sigma_c\) = 0.1 S/m and an intrinsic chargeability of 0.1 V/V. The resistor has a conductivity of \(\sigma_r\) = 0.001 S/m and is non-chargeable. Both blocks are oriented along the Northing direction and have x, y and z dimensions of 400 m, 1600 m and 320 m. Both blocks are buried at a depth of 160 m.
The data being inverted were generated using SimPEG. Synthetic apparent chargeability data were simulated with a pole-dipole configuration. The survey consisted of 9 West-East survey lines, each with a length of 2000 m. The line spacing was 250 m and the electrode spacing was 100 m. Gaussian noise with a standard deviation of 1e-6 V + 5% the absolute value were added to each datum. Uncertainties of 1e-6 V + 5% were assigned to the data for inversion.
SimPEG Package Details#
Link to the docstrings for the simulation class The docstrings will have a citation and show the integral equation.
Running the Inversion#
We begin by importing all necessary Python packages for running the notebook.
Show code cell source
from SimPEG.electromagnetics.static import induced_polarization as ip
from SimPEG.electromagnetics.static.utils.static_utils import (
plot_pseudosection,
plot_3d_pseudosection,
apparent_resistivity,
apparent_resistivity_from_voltage,
convert_survey_3d_to_2d_lines
)
from SimPEG.utils.io_utils import read_dcipoctree_ubc, write_dcipoctree_ubc
from SimPEG import maps, data, data_misfit, regularization, optimization, inverse_problem, inversion, directives
from discretize import TreeMesh
import matplotlib as mpl
import matplotlib.pyplot as plt
import plotly
import numpy as np
try:
from pymatsolver import Pardiso as Solver
except ImportError:
from SimPEG import SolverLU as Solver
mpl.rcParams.update({"font.size": 14})
write_output = True
A compressed folder containing the assets required to run the notebook is then downloaded. This includes the mesh, true model, and observed data files.
Show code cell source
# Import the .tar file
Extracted files are then loaded into the SimPEG framework.
Show code cell source
rootdir = './../../../assets/dcip/block_model_ip_inv_simpeg_octree/'
meshfile = rootdir + 'octree_mesh.txt'
chgfile = rootdir + 'true_model.chg'
backgroundfile = rootdir + 'background_cond.con'
dobsfile = rootdir + 'dobs_simpeg.txt'
mesh = TreeMesh.readUBC(meshfile)
true_chargeability_model = TreeMesh.readModelUBC(mesh, chgfile)
background_conductivity_model = TreeMesh.readModelUBC(mesh, backgroundfile)
ip_data = read_dcipoctree_ubc(dobsfile, 'apparent_chargeability')
Here, we plot the observed apparent chargeability data.
Show code cell source
survey = ip_data.survey
plane_points = []
for x in np.arange(-1000, 1100, 500):
p1, p2, p3 = np.array([-1000.,x,0]), np.array([1000,x,0]), np.array([1000,x,-1000])
plane_points.append([p1,p2,p3])
scene_camera=dict(
center=dict(x=-0.1, y=0, z=-0.2), eye=dict(x=1.2, y=-1, z=1.5)
)
scene = dict(
xaxis=dict(range=[-1000, 1000]), yaxis=dict(range=[-1000, 1000]), zaxis=dict(range=[-500, 0]),
aspectratio=dict(x=1, y=1, z=0.5)
)
vlim = [ip_data.dobs.min(), ip_data.dobs.max()]
fig2 = plot_3d_pseudosection(
ip_data.survey, ip_data.dobs, scale='lin', vlim=vlim,
plane_points=plane_points, plane_distance=10., units='V/V',
marker_opts={"colorscale": "plasma"}
)
fig2.update_layout(
title_text="Apparent Chargeability Data", title_x=0.5, width=800, height=700, scene_camera=scene_camera, scene=scene
)
plotly.io.show(fig2)
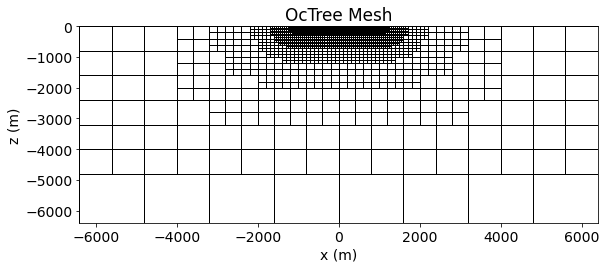
And here we plot the OcTree mesh used in the inversion. Since the topography is flat, all cells are active.
Show code cell source
ind = int(len(mesh.hy)/2)
fig = plt.figure(figsize=(9, 3.5))
ax1 = fig.add_axes([0.1, 0.12, 0.8, 0.78])
zeros_array = np.zeros(len(true_chargeability_model))
mesh.plot_slice(
zeros_array, ax=ax1, normal='Y', grid=True, ind=ind,
pcolorOpts={"cmap": "binary"}
)
ax1.set_title("OcTree Mesh")
ax1.set_xlabel("x (m)")
ax1.set_ylabel("z (m)")
Text(0, 0.5, 'z (m)')
Next, we define the mapping from the model space to the mesh and the simulation.
Show code cell source
eta_map = maps.IdentityMap()
simulation = ip.simulation.Simulation3DNodal(
survey=survey,
mesh=mesh,
etaMap=eta_map,
sigma=background_conductivity_model,
solver=Solver,
bc_type='Neumann'
)
We now define a starting model for the inversion. Assuming the background chargeability is negligible, we define a small starting model so a model update direction can be found on the first iteration.
Show code cell source
m0 = 1e-6 * np.ones(mesh.nC)
Here we define the measure of data misfit, the regularization and the algorithm used to compute the step-direction at each iteration. These are used to define the inverse problem.
Show code cell source
dmis = data_misfit.L2DataMisfit(data=ip_data, simulation=simulation)
reg_map = maps.IdentityMap(nP=mesh.nC)
reg = regularization.Simple(
mesh, mapping=reg_map,
alpha_s=0.1, alpha_x=1., alpha_y=1., alpha_z=1.
)
reg.mrefInSmooth = True
opt = optimization.ProjectedGNCG(
maxIter=10, maxIterLS=20, maxIterCG=30., tolCG=1e-2, lower=0.
)
inv_prob = inverse_problem.BaseInvProblem(dmis, reg, opt)
Here, we define the directives for the inversion.
Show code cell source
starting_beta = directives.BetaEstimate_ByEig(beta0_ratio=20.)
beta_schedule = directives.BetaSchedule(coolingFactor=2, coolingRate=2)
save_iteration = directives.SaveOutputEveryIteration(save_txt=False)
target_misfit = directives.TargetMisfit(chifact=1)
sensitivity_weights = directives.UpdateSensitivityWeights(truncation_factor=0.005)
update_jacobi = directives.UpdatePreconditioner()
directives_list = [
sensitivity_weights,
starting_beta,
beta_schedule,
save_iteration,
target_misfit,
update_jacobi
]
Finally, we define and run the inversion.
Show code cell source
inv = inversion.BaseInversion(inv_prob, directives_list)
simpeg_model = inv.run(m0)
simpeg_model = eta_map*simpeg_model
dpred = inv_prob.dpred
SimPEG.InvProblem will set Regularization.mref to m0.
SimPEG.InvProblem is setting bfgsH0 to the inverse of the eval2Deriv.
***Done using same Solver and solverOpts as the problem***
model has any nan: 0
=============================== Projected GNCG ===============================
# beta phi_d phi_m f |proj(x-g)-x| LS Comment
-----------------------------------------------------------------------------
x0 has any nan: 0
0 8.18e+05 7.45e+04 0.00e+00 7.45e+04 5.26e+04 0
1 8.18e+05 3.09e+03 6.77e-03 8.62e+03 4.27e+03 0
2 4.09e+05 2.80e+03 7.07e-03 5.68e+03 4.44e+03 0 Skip BFGS
3 4.09e+05 1.72e+03 8.90e-03 5.36e+03 1.11e+03 0
4 2.04e+05 1.73e+03 8.86e-03 3.54e+03 2.35e+03 0
5 2.04e+05 1.09e+03 1.11e-02 3.35e+03 3.81e+02 0
6 1.02e+05 1.10e+03 1.10e-02 2.22e+03 1.80e+03 0
7 1.02e+05 6.99e+02 1.38e-02 2.11e+03 2.79e+02 0
8 5.11e+04 7.02e+02 1.37e-02 1.40e+03 1.42e+03 0
------------------------- STOP! -------------------------
1 : |fc-fOld| = 0.0000e+00 <= tolF*(1+|f0|) = 7.4476e+03
0 : |xc-x_last| = 6.0887e-01 <= tolX*(1+|x0|) = 1.0004e-01
0 : |proj(x-g)-x| = 1.4181e+03 <= tolG = 1.0000e-01
0 : |proj(x-g)-x| = 1.4181e+03 <= 1e3*eps = 1.0000e-02
0 : maxIter = 10 <= iter = 9
------------------------- DONE! -------------------------
If desired, we can output the recovered model and the predicted data.
Show code cell source
if write_output:
TreeMesh.writeModelUBC(mesh, rootdir+'recovered_model.chg', simpeg_model)
data_dpred = data.Data(survey=ip_data.survey, dobs=dpred)
write_dcipoctree_ubc(rootdir+'dpred.txt', data_dpred, 'apparent_chargeability', 'dpred')
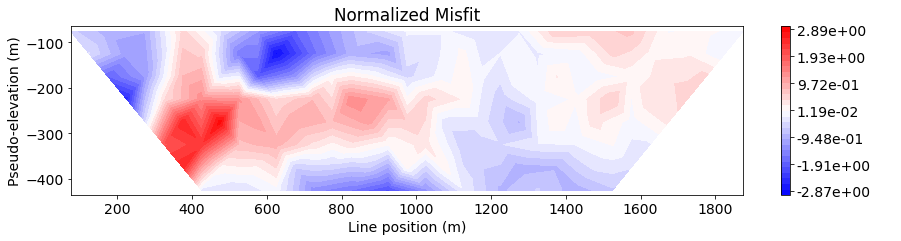
Data Misfit Along a Single Survey Line#
Here, we examine the data misfit by plotting a 2D pseudo-section for each survey line. We begin by parsing the 3D survey into a list of 2D surveys.
Show code cell source
line_id = np.zeros(survey.nD)
northing = np.unique(survey.locations_a[:, 1])
for ii in range(0, len(northing)):
line_id[survey.locations_a[:, 1] == northing[ii]] = ii
survey_2d_list = convert_survey_3d_to_2d_lines(survey, line_id, 'apparent_chargeability')
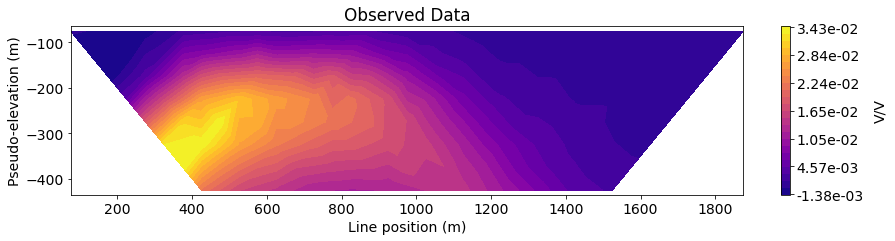
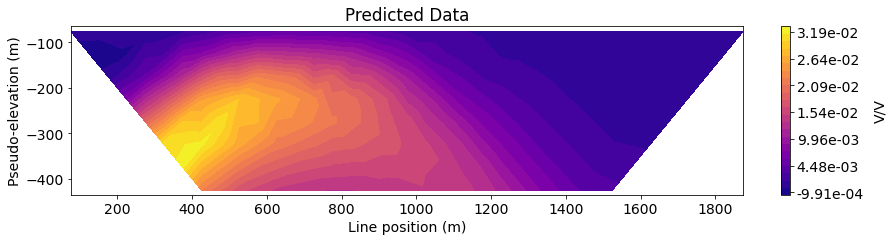
Next, we define the line ID (0-8) for the pole-dipole line we would like to examine. Then plot the observed data, predicted data and normalized misfit in 2D pseudosection.
Show code cell source
IND = 4
title_str = ['Observed Data', 'Predicted Data', 'Normalized Misfit']
data_list_2d = [
ip_data.dobs,
dpred,
(ip_data.dobs - dpred) / ip_data.standard_deviation
]
cmap_list = [mpl.cm.plasma, mpl.cm.plasma, mpl.cm.bwr]
label_list = ["V/V", "V/V", " "]
for ii in range(0, 3):
fig = plt.figure(figsize=(14, 3))
ax1 = fig.add_axes([0.1, 0.15, 0.75, 0.78])
plot_pseudosection(
survey_2d_list[IND],
dobs=data_list_2d[ii][line_id==IND],
plot_type="contourf",
ax=ax1,
vlim=vlim,
cbar_label=label_list[ii],
contourf_opts={"levels": 30, "cmap": cmap_list[ii]},
)
ax1.set_title(title_str[ii])
plt.show()
D:\Documents\Repositories\simpeg\SimPEG\electromagnetics\static\utils\static_utils.py:578: UserWarning:
plot_pseudosection unused kwargs: {list(kwargs.keys())}
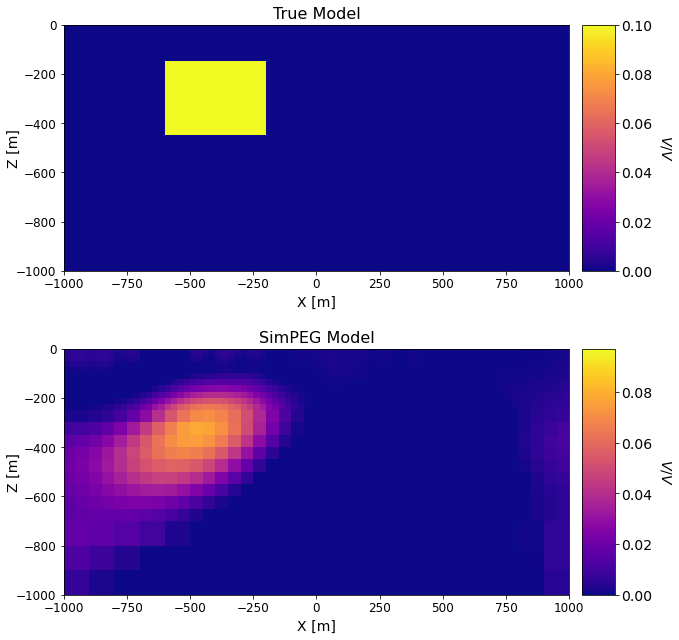
Comparing True and Recovered Models#
Show code cell source
fig = plt.figure(figsize=(9, 9))
font_size = 14
models_list = [true_chargeability_model, simpeg_model]
titles_list = ['True Model', 'SimPEG Model']
ax1 = 2*[None]
cplot = 2*[None]
ax2 = 2*[None]
cbar = 2*[None]
for qq in range(0, 2):
ax1[qq] = fig.add_axes([0.1, 0.55 - 0.5*qq, 0.78, 0.38])
cplot[qq] = mesh.plot_slice(
models_list[qq], normal='Y', ind=int(len(mesh.hy)/2),
grid=False, ax=ax1[qq], pcolorOpts={"cmap": "plasma"}
)
cplot[qq][0].set_clim((np.min(models_list[qq]), np.max(models_list[qq])))
ax1[qq].set_xlim([-1000, 1000])
ax1[qq].set_ylim([-1000, 0])
ax1[qq].set_xlabel("X [m]", fontsize=font_size)
ax1[qq].set_ylabel("Z [m]", fontsize=font_size, labelpad=-5)
ax1[qq].tick_params(labelsize=font_size - 2)
ax1[qq].set_title(titles_list[qq], fontsize=font_size + 2)
ax2[qq] = fig.add_axes([0.9, 0.55 - 0.5*qq, 0.05, 0.38])
norm = mpl.colors.Normalize(vmin=np.min(models_list[qq]), vmax=np.max(models_list[qq]))
cbar[qq] = mpl.colorbar.ColorbarBase(
ax2[qq], norm=norm, orientation="vertical", cmap=mpl.cm.plasma
)
cbar[qq].set_label(
"$V/V$", rotation=270, labelpad=20, size=font_size,
)
plt.show()